Results

A variety of different experiments were performed to determine the strengths and limitations of our system. Additional
effort was focused on improving the squared error and handling target movement during the collection of the
projections. Each of the following sections addresses the results of these efforts.
In all of the following images, any speckle noise is the result of artifacts during the conversion from MATLAB's image
display to the GIF image file format used on these pages. A lack of available colors resident in the
computer's graphics table, image scaling, and the file format itself produces these artifacts.
Case I
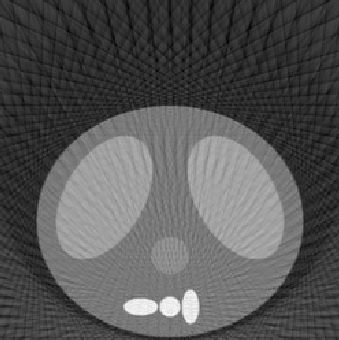
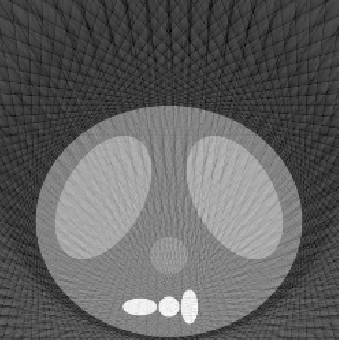
The first experiment was to determine the performance of our algorithm under three variables: number of projection angles, detector length, and number of detectors. The following
reconstructed images represent a good sampling to demonstrate the strengths and weaknesses of the methodology. The squared error (SE) is also listed to give a quantitative comparison, along with the qualitative differences shown in the reconstructed images. The phantom image from which the reconstructions were made is shown at the top of Fig. 5.1. All reconstructed images for this experiment used the fftfilter.m in the backproj.m function.

Number of Projection Angles = 45
| Number of Detectors on the Detector Array |
| 125 | 250 | 500 | |
| Detector Array Length 0.50 |
 |
 |
 |
| SE = 11092 | SE = 16776 | SE = 26032 | |
| 1.00 | 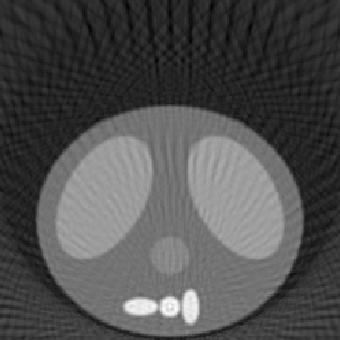 |
 |
 |
| SE = 205.3 | SE = 231.9 | SE = 286.9 | |
| 1.50 | 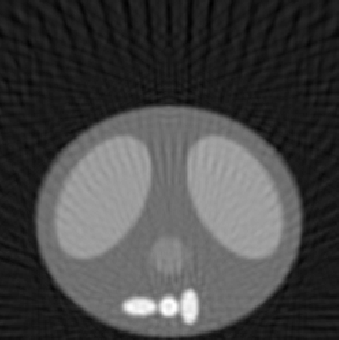 |
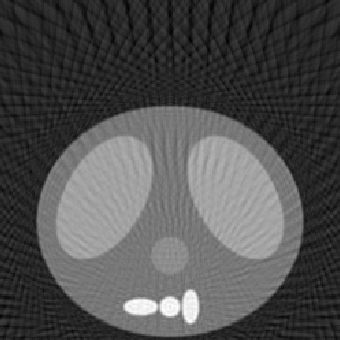 |
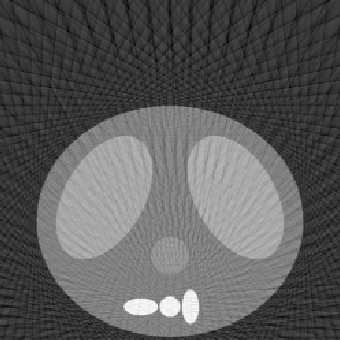 |
| SE = 94.1 | SE = 110.7 | SE = 158.2 |
Number of Projection Angles = 135
| Number of Detectors on the Detector Array |
| 125 | 250 | 500 | |
| Detector Array Length 0.50 |
 |
 |
 |
| SE = 9810 | SE = 13853 | SE = 19743 | |
| 1.00 | 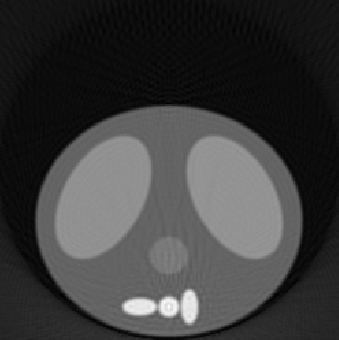 |
 |
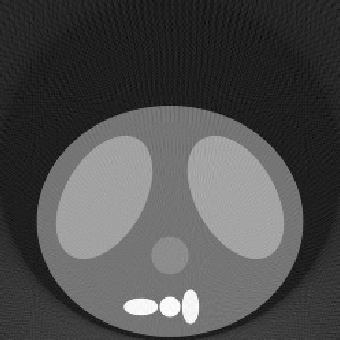 |
| SE = 153.2 | SE = 129.5 | SE = 128.8 | |
| 1.50 |  |
 |
 |
| SE = 65.8 | SE = 36.4 | SE = 31.3 |
Number of Projection Angles = 250
| Number of Detectors on the Detector Array |
| 125 | 250 | 500 | |
| Detector Array Length 0.50 |
 |
 |
 |
| SE = 9589 | SE = 13294 | SE = 18480 | |
| 1.00 |  |
 |
 |
| SE = 151.2 | SE = 121.3 | SE = 109.5 | |
| 1.50 |  |
 |
 |
| SE = 65.5 | SE = 32.9 | SE = 18.5 |
Number of Projection Angles = 400
| Number of Detectors on the Detector Array |
| 125 | 250 | 500 | |
| Detector Array Length 0.50 |
 |
 |
 |
| SE = 9575 | SE = 13122 | SE = 17932 | |
| 1.00 |  |
 |
 |
| SE = 151.2 | SE = 120.3 | SE = 105.5 | |
| 1.50 |  |
 |
 |
| SE = 65.5 | SE = 32.7 | SE = 16.5 |
algorithm using the labeled number of projection angles, detector
length, and number of detectors. Squared error (SE) is also listed.
Case II
First inspection of the reconstructed images noted that some parts of the background have negative values. Although most of these values were small, this implies that a detector could receive a negative quantity of x-rays. Logically this does not make sense. The cause of the negative values comes from computational roundoff, and Gibb's phenomenon in the discrete Fourier transform filtering (fftfilter.m).
Removal of the negative values is reasonable since the difference between the background and any actual mass between the source and detectors is relatively large. An effort to remove the negative values should not disturb any real structure we want to observe. A simple program, zeroneg.m, was used to change all negative values to zero. The results are shown in Fig. 5.2.
| Before | After | |
| Number of Angles = 250 Length of Detector = 1.5 No. of Detectors = 500 |
 |
 |
| SE = 18.5 | SE = 17.1 | |
| Number of Angles = 400 Length of Detector = 1.5 No. of Detectors = 500 |
 |
 |
| SE = 16.5 | SE = 16.0 |
before and after changing all negative values to zero. Squared error
(SE) is also listed.
Case III
In this experiment, reconstructed images similar to Case I were obtained using the convolution back-projection algorithm (i.e., using convfilter.m instead of fftfilter.m).
Number of Projection Angles = 400
| Number of Detectors on the Detector Array |
| 125 | 250 | 500 | |
| Detector Array Length 0.50 |
 |
 |
 |
| SE = 7343 (9575) | SE = 8376 (13122) | SE = 10471 (17932) | |
| 1.0 |  |
 |
 |
| SE = 39.59 (151.2) | SE = 19.45 (120.3) | SE = 11.47 (105.5) | |
| 1.50 |  |
 |
 |
| SE = 60.2 (65.5) | SE = 29.3 (32.7) | SE = 15.26 (16.5) |
instead of the FFT based reconstruction algorithm using the labeled
number of projection angles, detector length, and number of
detectors. Squared error is also listed, along with the squared
error from Case I shown in ().
The above result shows an interesting difference in the two
implementations of the filter. It can be seen that when the
convolution based filter is used, the performance for detector length
1.0 is the best. This is possible because:
1. The sampling density of the projection is larger in this case than for a detector length of
1.5.
2. The non-zero portion of the thoracic cavity phantom is within the detector array.
The same is not observed, however, when the FFT based filter is
used. This is due to the following difference in their
implementation. In the FFT based implementation, the filtered
projection is restricted to the same length as the projection. In the
convolution based implementation, the filtered projection is longer
than the projection. Due to this difference, the portions of the image
that are not within the detector (the areas near the four corners of
the image) are not reconstructed properly in the FFT based
implementation but reconstructed well in the convolution based
implementation. The top two corners are reconstructed well in both
methods since most of the back-projection values for them will be
zero. However, the bottom two corners contribute to most of the error
in the FFT based implementation.
The following squared error images illustrate this.
| FFT based filter | Convolution based filter | ||
| Detector Array Length = 1.0 Number of Detectors = 500 Number of projection angles = 400 |
 |
 |
|
| SE = 105.5 | SE = 11.47 |
Total squared error (SE) is also listed.
Case IV
In this experiment, we simulate the effect of shifts in the entire image and growth in specific regions like the lungs and heart. These changes occur during the process of determining the projections. In the specific simulation results shown, the shift, grow_lng or grow_hrt parameter used in project.m is modified halfway through the process of determining the projections.
| Shift in Image | Growth of Heart | Growth of Lung |
 |
 |
 |
| SE = 433.79 | SE = 19.86 | SE = 573.26 |
or growth of regions, occurring halfway through the process of
determining the projections. Number of projection angles = 400.
Number of detectors = 500. Detector length = 1.5. The shift and
growth parameters were changed after 200 projections were
obtained.
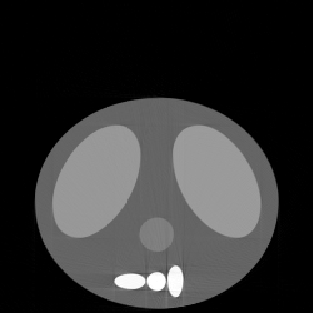
| Shift in Image | ||
| with Correction | ||
 |
||
| SE = 36.88 |
during the collection of projections (using shiftfix.m). Note the artifacts
near the spinal column.
Case V
This section contains the results from our attempts at image enhancement. Six images were used and are listed below in Table 5.1. All reconstructed images for this experiment used the fftfilter.m in the backproj.m function. Each row corresponds to a different filter, described in the image enhancement section. The metric used to determine image quality was the total squared error between each reconstructed and filtered pixel and the corresponding original image pixels.
Three things are immediately obvious from the above table. The first is that
most of the squared error in images 2 and 3, which have low projection angles
and detector array quantities compared to image 1, can be eliminated with
the thresholding filter. Therefore, the biggest problem with smaller detector
and projection amounts is the introduction of low valued artifacts outside
the cavity. Secondly, we see that we were not able to improve much on
reconstructed images where the lungs were expanded or the phantom shifted.
Finally, we see that the edge thinning through the Haar wavelet transform
actually increased the squared error. However, while the metric was
increased, we see in the figures below that the edges are crisper.
| Before | After | ||
 |
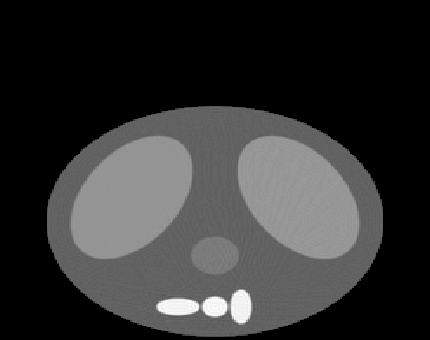 |
wavelet transform edge thinning algorithm.
Commentary on these results can be found in our
conclusions section.
Final Results
After looking at the variety of outcomes from the above experiments, we applied our best algorithms to a single reconstruction. The final reconstructed image was generated using 400 projection angles, a length one detector, and 500 detectors on the array. We used the convolution back-projection to filter the projections. After the image was formed, post-processing algorithms included both zeroneg.m and median filtering. The final squared error was 9.61.
| Original Thoracic Image | Final Reconstruction |
 |
 |
| Image of Squared Error | 3-D Image of Squared Error |
 |
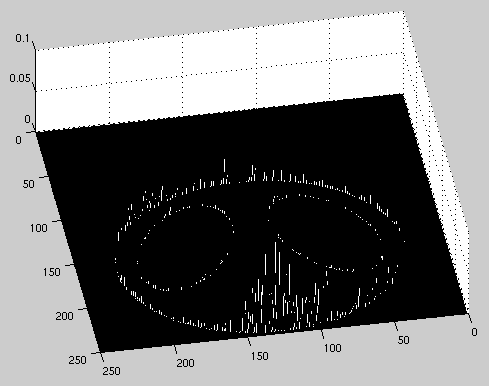 |
along with plots of the error locations.